- by Jimmy Fisher
- Nov 02, 2024
Wrangling BRFSS (2011-2023)
- By Jimmy Fisher
- Dec 01, 2024
- in Coding Projects

The Behavioral Risk Factor surveillance System (BRFSS) is a health-related telephone survey established in 1984 that gathers data on health risk behaviors, chronic conditions, and preventive service use among U.S. residents. It now covers all 50 states and includes 400,000 adult interviews annually, making it the largest ongoing health survey system globally.
So, it's worth checking out, though dealing with it outside of the provided SAS tables and code can sometimes get a little tedious. Several base data tables can be downloaded from cdc.gov HERE, though I have converted the full BRFSS annual data deliveries from 2011 to 2023 into separate .csv files, for your convenience:
- BRFSS 2011 (450 variables)
- BRFSS 2012 (354 variables)
- BRFSS 2013 (330 variables)
- BRFSS 2014 (275 variables)
- BRFSS 2015 (330 variables)
- BRFSS 2016 (275 variables)
- BRFSS 2017 (358 variables)
- BRFSS 2018 (275 variables)
- BRFSS 2019 (342 variables)
- BRFSS 2020 (279 variables)
- BRFSS 2021 (303 variables)
- BRFSS 2022 (326 variables)
- BRFSS 2023 (350 variables)
In all of these data files, I have added an 'x' to the beginning of every column name beginning with an underscore since R doesn't let variable names begin with underscores. So, for example, in the 2023 data dictionary, the variable _PASTRNG is a variable indicating whether respondents met the muscle strengthening recommendations. In the above .csv file for 2023, this variable will import as x_PASTRNG.
Also, as you may already have experienced, and/or will glimpse if you search the web for handling BRFSS data in R or Python, the variable encoding is its own adventure. Some variables appear or vanish across years, and others have their encoding slightly altered. For example, the 2023 codebook clarifies that a value of '10' in the IMFVPLA4 column means that the respondent's last flu vaccine was received in Canada or Mexico, whereas a value of '10' in 2022's analogous IMFVPLA3 variable means, according the 2022 codebook, that the respondent's last flu vaccine was received at a school. In 2023, an '11' rather than a '10' indicates last flu shot receipt at a school, as "Received vaccination in Canada/Mexico" was newly added to the variable in 2023. One good thing: When there is a change to any variable encoding, its name gets incremented (e.g., IMFVPLA3 in 2022 to IMFVPLA4 in 2023).
To make things easier for you, I created a value labels map for the encoded variables within every full BRFSS annual data file from 2011 to 2023 and provide it here as an RDS file. It'll get you 98% of the way there, and I've added a beginning 'x' to every computed variable (beginning with an underscore) to match the above .csv files. So, after importing BRFSS_Full_350_2023.csv into R (from the above list), you could use the .rds file to replace encoding for specified variables, in the below example for SEXVAR and x_PASTRNG.
# Load 2023 BRFSS datadata <- read.csv("BRFSS_Full_350_2023.csv")
# Load the map from value_label_map.rdsvalue_label_map <- readRDS("BRFSS_label_map_2011-2023.rds")
## Map data variables selectively using array of columnsvariables_to_map <- c("SEXVAR", "x_PASTRNG")for (var in variables_to_map) { # Use the value_label_map to replace values in the data data[[var]] <- value_label_map[[var]][as.character(data[[var]])]}
# Load 2023 BRFSS data
data <- read.csv("BRFSS_Full_350_2023.csv")
# Load the map from value_label_map.rds
value_label_map <- readRDS("BRFSS_label_map_2011-2023.rds")
## Map data variables selectively using array of columns
variables_to_map <- c("SEXVAR", "x_PASTRNG")
for (var in variables_to_map) {
# Use the value_label_map to replace values in the data
data[[var]] <- value_label_map[[var]][as.character(data[[var]])]
}
This replaces all of the numbers in each of the specified columns with the meaning of those numbers (e.g., for SEXVAR, '1' is Male and '2' is Female).
You can also print values directly from the .rds file. However, since variables in R cannot begin with underscores, x_RFSEAT3:
# Print values for variable _RFSEAT3head(value_label_map[["x_RFSEAT3"]])
# Print values for variable _RFSEAT3
head(value_label_map[["x_RFSEAT3"]])
Finally, if you wanted to, you could replace all encoded values in a year of imported BRFSS data by iterating through the .rds like this:
variables_to_map <- intersect(names(data), names(value_label_map))for (var in variables_to_map) { # Use the value_label_map to replace values in the data data[[var]] <- value_label_map[[var]][as.character(data[[var]])]}
variables_to_map <- intersect(names(data), names(value_label_map))
for (var in variables_to_map) {
# Use the value_label_map to replace values in the data
data[[var]] <- value_label_map[[var]][as.character(data[[var]])]
}
While it makes sense to leave the the variables encoded during analysis, at any point this approach can be used to translate BRFSS data variable values into their meanings.
One thing that we might want to do with these survey data is compare trends across years. To do this, we need to map what variables are tracked across multiple years. We can save a lot of time by using R to delimit a variable pool for possible cross-year comparison.
# Import dataall_data <- data.frame(year = character(), variable = character(), stringsAsFactors = FALSE)
# Define the data filesdata_files <- c("./data/2011_450.csv", "./data/2012_354.csv", "./data/2013_330.csv", "./data/2014_275.csv", "./data/2015_330.csv", "./data/2016_275.csv", "./data/2017_358.csv", "./data/2018_275.csv", "./data/2019_342.csv", "./data/2020_279.csv", "./data/2021_303.csv", "./data/2022_326.csv", "./data/2023_350.csv")
# Pull out the year and variable names from each filefor (file in data_files) { year <- substr(basename(file), 1, 4) data <- read.csv(file) variables <- colnames(data) temp_data <- data.frame( year = rep(year, length(variables)), variable = variables, stringsAsFactors = FALSE ) all_data <- rbind(all_data, temp_data)}
head(all_data)
# Create a dataframe with every unique variable in all_dataunique_vars <- sort(unique(all_data$variable))years <- 2011:2023
# Initialize the dataframevars <- data.frame(variable = unique_vars, stringsAsFactors = FALSE)
# Add columns for each yearfor (year in years) { vars[[as.character(year)]] <- ""}
# Populate the dataframefor (i in 1:nrow(vars)) { var <- vars$variable[i] for (year in years) { if (any(all_data$variable == var & all_data$year == as.character(year))) { vars[i, as.character(year)] <- "x" } }}
head(vars)
# Save the dataframe to a CSV filewrite.csv(vars, "variable_coverage.csv", row.names = FALSE)
# Import data
all_data <- data.frame(year = character(), variable = character(), stringsAsFactors = FALSE)
# Define the data files
data_files <- c("./data/2011_450.csv",
"./data/2012_354.csv",
"./data/2013_330.csv",
"./data/2014_275.csv",
"./data/2015_330.csv",
"./data/2016_275.csv",
"./data/2017_358.csv",
"./data/2018_275.csv",
"./data/2019_342.csv",
"./data/2020_279.csv",
"./data/2021_303.csv",
"./data/2022_326.csv",
"./data/2023_350.csv")
# Pull out the year and variable names from each file
for (file in data_files) {
year <- substr(basename(file), 1, 4)
data <- read.csv(file)
variables <- colnames(data)
temp_data <- data.frame(
year = rep(year, length(variables)),
variable = variables,
stringsAsFactors = FALSE
)
all_data <- rbind(all_data, temp_data)
}
head(all_data)
# Create a dataframe with every unique variable in all_data
unique_vars <- sort(unique(all_data$variable))
years <- 2011:2023
# Initialize the dataframe
vars <- data.frame(variable = unique_vars, stringsAsFactors = FALSE)
# Add columns for each year
for (year in years) {
vars[[as.character(year)]] <- ""
}
# Populate the dataframe
for (i in 1:nrow(vars)) {
var <- vars$variable[i]
for (year in years) {
if (any(all_data$variable == var & all_data$year == as.character(year))) {
vars[i, as.character(year)] <- "x"
}
}
}
head(vars)
# Save the dataframe to a CSV file
write.csv(vars, "variable_coverage.csv", row.names = FALSE)
The output file, variable_coverage.csv provides a variable mesh across all years of the above BRFSS data, 2011-2023. Think of this in at least 4 segments:
1) Direct cross-comparisons across all 13 years of provided data, because the same encoded variables were provided every year. From 2011-2023, the 13 years of BRFSS data I provided above as .csv files, 71 variables remain the same across all files.
2) Direct cross-comparisons across some years. For example, if we constrain our time horizon to 2019-2023, 162 variables have remained the same.
3) Indirect cross-comparisons across years following slight alterations to some variables. For example, COVIDNU1 in 2022 and COVIDNU2 in 2023 both capture responses to the same question: "How many COVID-19 vaccinations have you received?" However, whereas a '4' in COVIDNU1 meant "Four or more" in 2022, a '4' in 2023's COVIDNU2 means "four" because a '5' means "Five or more" vaccines were received. Since there was no '5' in COVIDNU1 from 2022, there is no direct way to map these variables across years. However, technically, a data scientist could recode both '4' and '5' in as '4' and designate it as "Four or more," making responses indirectly comparable across years. There are reasons not to do this, but it is worth pointing out that some changed variables could theoretically be mapped across years for indirect comparison.
4) Variables whose capture or derivation is either completely missing across years or else has changed so completely as to make any comparisons dubious. One cannot analyze data that do not exist.
For fun, let's start by exploring the 162 variables that have remained the same from 2019 to 2023. I pulled these by defining a vector array of variables (vars_of_interest) and pulling them into a single dataframe from each year's separate .csv file. I added a 'year' column to designate where data were coming from and, after combining these dataframes, saved it as a single .csv file, brfss_162_2019to2023.csv.
# Import datavars_of_interest <- read.csv("variable_coverage.csv")
# Keep variables that have an x in 2019-2023vars_of_interest <- vars_of_interest[rowSums(vars_of_interest[, 10:14] == "x") > 4, ]
# Save variable column values to a listvars_of_interest <- as.vector(vars_of_interest$variable)
# Load vars_of_interest from 2019-2023 BRFSS databrfss_162_2019 <- read.csv("./data/2019_342.csv")[vars_of_interest]brfss_162_2020 <- read.csv("./data/2020_279.csv")[vars_of_interest]brfss_162_2021 <- read.csv("./data/2021_303.csv")[vars_of_interest]brfss_162_2022 <- read.csv("./data/2022_326.csv")[vars_of_interest]brfss_162_2023 <- read.csv("./data/2023_350.csv")[vars_of_interest]
# Add a year column to each dataframebrfss_162_2019$year <- 2019brfss_162_2020$year <- 2020brfss_162_2021$year <- 2021brfss_162_2022$year <- 2022brfss_162_2023$year <- 2023
# Combine the dataframesbrfss_162_2019to2023 <- rbind(brfss_162_2019, brfss_162_2020, brfss_162_2021, brfss_162_2022, brfss_162_2023)
# Save the combined dataframe to a CSV filewrite.csv(brfss_162_2019to2023, "brfss_162_2019to2023.csv", row.names = FALSE)
# Import data
vars_of_interest <- read.csv("variable_coverage.csv")
# Keep variables that have an x in 2019-2023
vars_of_interest <- vars_of_interest[rowSums(vars_of_interest[, 10:14] == "x") > 4, ]
# Save variable column values to a list
vars_of_interest <- as.vector(vars_of_interest$variable)
# Load vars_of_interest from 2019-2023 BRFSS data
brfss_162_2019 <- read.csv("./data/2019_342.csv")[vars_of_interest]
brfss_162_2020 <- read.csv("./data/2020_279.csv")[vars_of_interest]
brfss_162_2021 <- read.csv("./data/2021_303.csv")[vars_of_interest]
brfss_162_2022 <- read.csv("./data/2022_326.csv")[vars_of_interest]
brfss_162_2023 <- read.csv("./data/2023_350.csv")[vars_of_interest]
# Add a year column to each dataframe
brfss_162_2019$year <- 2019
brfss_162_2020$year <- 2020
brfss_162_2021$year <- 2021
brfss_162_2022$year <- 2022
brfss_162_2023$year <- 2023
# Combine the dataframes
brfss_162_2019to2023 <- rbind(brfss_162_2019, brfss_162_2020, brfss_162_2021, brfss_162_2022, brfss_162_2023)
# Save the combined dataframe to a CSV file
write.csv(brfss_162_2019to2023, "brfss_162_2019to2023.csv", row.names = FALSE)
Variable encodings can be handled using the above .rds file, but what survey question is associated with each column name? For this, I visited the 2023 BRFSS documentation and pulled each variable's question into this spreadsheet.
In cross-referencing our 162 variables with this data dictionary, you'll notice a few things. First, several variables have only "HIDDEN" values, used to determine weights of each record, their contribution to generalizable data observations. Other variables are essentially meaningless to population characteristics, such as the day and month on which the survey was administered, and multiple confirmatory questions on whether a phone number is a landline.
Finally, several variables redundantly overlap. Age is cut into various category sizes, including 2, 6, and 14-level age variables. Respondent height is reported in both imperial and metric units, and BMI is provided as a raw value, an overweight/obese flag, and a 4-category BMI designation. Some imputed variables combine other columns, such as adults aged 65+ who have ever had a pneumonia vaccination (x_PNEUMO3).
Here are the variables that I picked out to compare from pre-COVID 2019 to post-COVID 2023, with highlighted respondent demographic variables to compare between:
The highlighted variables constituting the group differences I would like to compare between are age (x_AGEG5YR), sex (SEXVAR), educational attainment (x_EDUCAG), state of residence (x_STATE), and whether the respondents live in an urban or rural environment (x_URBSTAT).
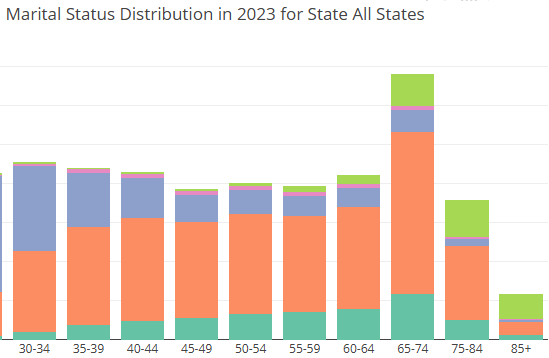
For my purposes here, I built a few simple visualizations wrapped within an R Shiny app to compare proportions of how some categorical survey answers changed across each combination of these 5 respondent groups. The end result was an R Shiny App that can assist with hypothesis generation.
Code (on GitHub) <- includes data prep
Deployment
To be clear, this exploratory data analysis (EDA) approach contains neither statistical methods nor sufficient detail to determine statistically significant change or differences across groups. This is also all self-reported data... But, it's a start!
For those who are interested, below are a couple examples of statistical tests that can be run relatively easily on BRFSS data. If there isn't a hyperlink attached yet, then it is planned for the future but not yet carried out.
- Chi Square Tests to assess whether a variable has statistically significantly changed over time (and how to use a post hoc Bonferroni test to determine which categories have changed)
- Multivariate Linear Regression to predict self-reported mental health